Circulating glycoprotein acetyls (GlycA) is a nuclear magnetic resonance (NMR) spectroscopy signal that reflects a composite or overall measure of changes in both, the number and the complexity of N-glycan side chains attached to acute phase reactant proteins (e.g., fibrinogen and C-reactive protein) under states of both acute and chronic inflammation, and therefore have potential in providing insights into disease activity and pathophysiological process. GlycA has been gaining increased interest as a serum biomarker for systemic inflammation and has been found to be useful in improving risk prediction for various diseases, including types of cardiovascular diseases, cancer, and all-cause mortality. However, the associations of GlycA with lung function and respiratory diseases remain to be investigated. Prokić et al. found that higher levels of plasma GlycA were significantly associated with COPD, and further found that GlycA was a biomarker of inflammatory pathways in the early stage of COPD in a prospective cohort. However, it is unclear the underlying mechanisms and if there is causality in explaining the above observed associations.

Recently, a study by Weihong Chen and Yanjun Yanjun Guo’s team identified significant whole-genome genetic correlations of GlycA with lung function parameters, asthma, and COPD, and consistently identified ten shared loci (including chr3p21.31 and chr8p23.1) at both SNP and gene level revealing potential shared biological mechanisms involving ubiquitination, immune response,Wnt/β-catenin signaling, cell growth and differentiation in tissues or cells including blood, epithelium, fibroblast, fetal thymus, and fetal intestine. The findings provide new genetic evidence for understanding the role of GlycA in respiratory diseases and highlight its potential therapeutic value for improving lung function and treating respiratory diseases.The study, titled “Genetic Association of Inflammatory Marker GlycA with Lung Function and Respiratory Diseases”, was published in Nature Communications.
In this study, the authors conducted extensive bioinformatics analyses using large-scale genomic data, including those from the UK Biobank and FinnGen. As illustrated in Figure 1, cross-trait linkage disequilibrium score regression analyses revealed significant genetic correlations between GlycA and lung function parameters (FEV1 and FVC), as well as asthma and COPD, elucidating a shared genetic architecture. To further investigate whether the observed whole-genome genetic correlation of GlycA with lung function parameters, asthma, and COPD are attributed to specific functional regions, they estimated annotation specific genetic correlations of GlycA with lung function parameters, asthma, and COPD in 11 large annotations using partitioned LDSC. Significant partitioned genetic correlation of GlycA with FEV1, FVC, asthma and COPD was observed within DGF, DHS, fetal DHS, TFBS, and transcribed region (Figure 2).
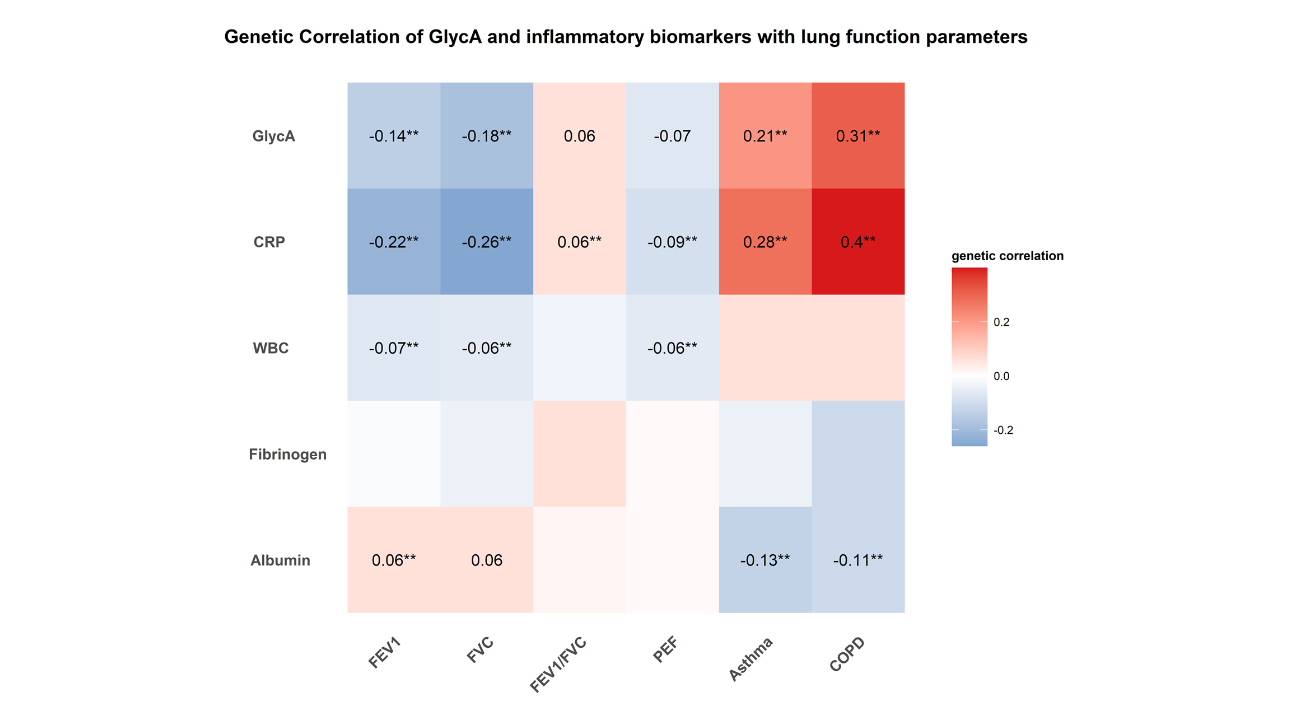
Figure 1 Genetic correlations of GlycA and other systemic inflammatory markers with lung function, asthma and COPD
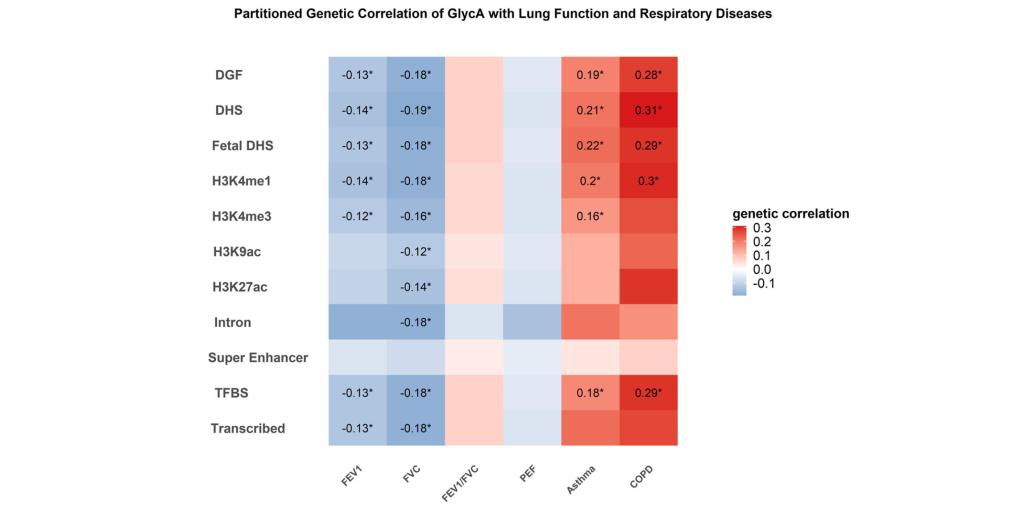
Figure 2 Partitioned genetic correlations of GlycA with lung function, asthma and COPD according to 11 functional categories
To further explore the existence of shared genetic variants that may underlie the significant genetic correlation observed in the LDSC analysis of GlycA with lung function or respiratory diseases, cross-trait meta-analysis and TWAS analysis were conducted to identify associated shared genetic loci from the SNP level and gene level, respectively. At the SNP level, regions such as chr3p21.31 and chr19q13.32 were identified. At the gene level, loci including UBA7 (located at chr3p21.31) and AFF1 (located at chr4q21.3) were identified. Notably, ten of these shared TWAS genes (chr1q21.3, chr1q41, chr3p21.31, chr4p16.3, chr4q21.3, chr5q14.1, chr5q31.1, chr8p23.1, chr9q34.2, and chr10q21.3) were also identified from SNP-level cross-trait meta-analysis. To investigate the biological pathways of the shared genes from cross-trait meta-analysis, GO and KEGG pathway enrichment analyses were conducted to assess the enrichment of shared genes of GlycA with lung function parameters and respiratory diseases. Additionally, the relative enrichment of the shared signals for GlycA with lung function parameters and respiratory diseases in different cell types was assessed, showing predominant enrichment in blood, epithelium, fibroblast, fetal thymus, fetal spleen, and fetal intestine tissues (Figure 3), consistent with the pathway analysis results. Finally, bidirectional Mendelian randomization analysis provided strong evidence for the causal association between GlycA and lung function parameters and respiratory diseases.
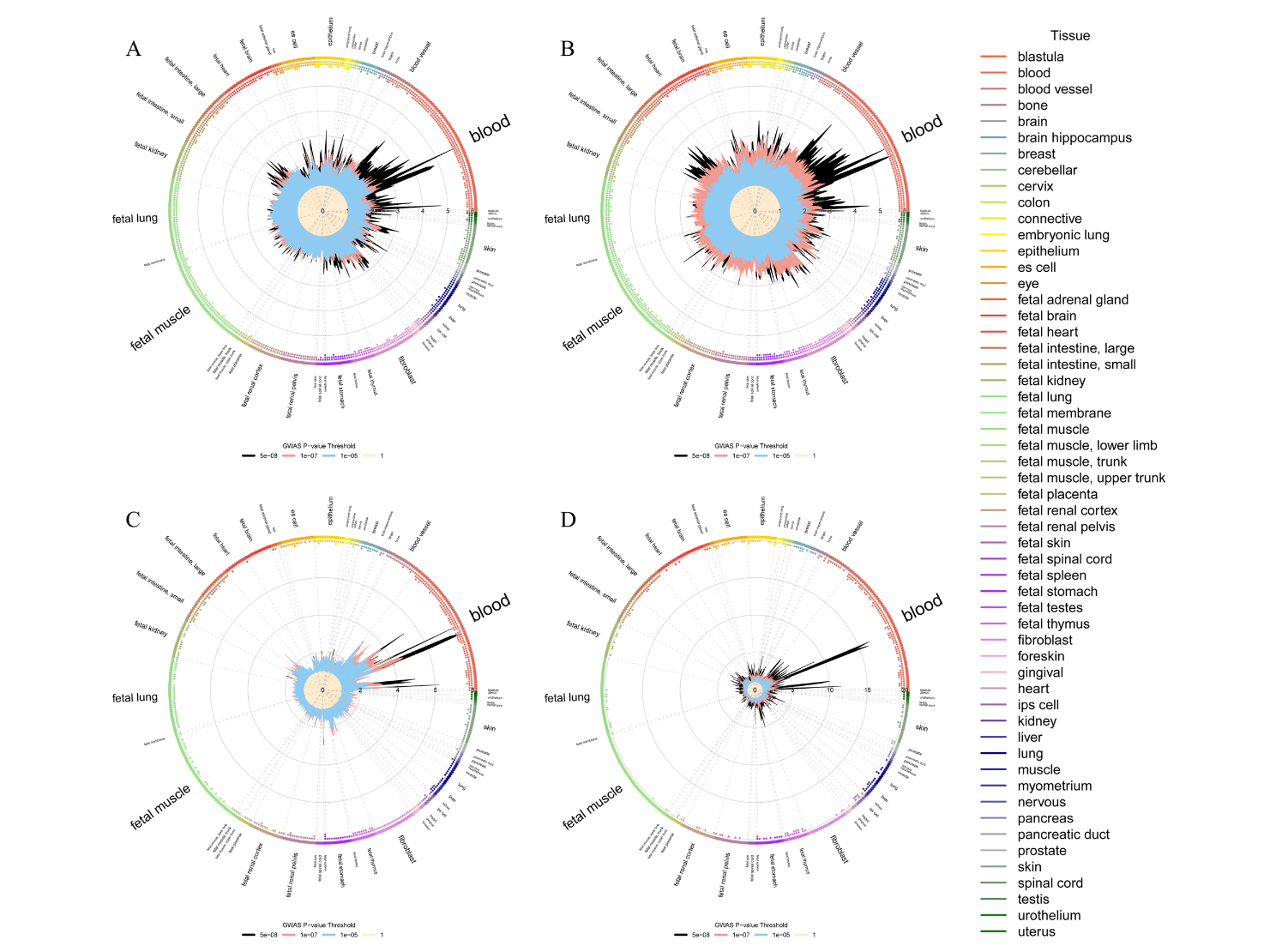
Fig. 3 Relative enrichment of shared signals in different cell types
In summary, the findings further our understanding of the role of GlycA in lung function, asthma, and COPD susceptibility, reveal the prominent genetic-based role and potential shared mechanisms by identifying significant instrumental effects and shared genetic components including IL6R, UBA7, CTSB, SOX7,andTOMM40-APOE-APOC1, all of which may provide insights into biological mechanisms of lung function, asthma, and COPD.

